
Programmable
DNA debuts
By
Kimberly Patch,
Technology Research NewsThe DNA molecule is the mechanism nature uses to construct every living being. Scientists are beginning to use the same mechanism to compute, but most of the rudimentary DNA computers built so far require researchers to manually trigger each step of a molecular calculation.
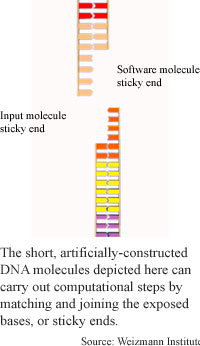
Researchers from the Weizmann Institute in Israel have found a way to automatically carry out several computational steps at once in a test tube of short, artificially-constructed DNA molecules. They put the DNA through a series of steps that, once begun, carried through to the end of a computation.
Although it takes minutes for DNA to do a single computation, many molecules can compute at once in a very small amount of space. "We get a trillion computers doing a billion operations per second in [a drop] of solution," said Ehud Shapiro, an associate professor in the departments of computer science and biological chemistry at the Weizmann Institute of Science in Israel.
This computation DNA, like biological DNA, is made up of four types of bases attached to long sugar-phosphate chains. Two of these chains, with bases attached, zip together, pairing up the bases on each chain to form DNA's classic double-helix shape.
Biological DNA uses specific sequences of the bases as blueprints to build the many proteins involved in the chemical reactions of life. The same sequences can represent numbers and be manipulated mathematically to compute.
The computation method uses two types of DNA molecules: software, or computation molecules, which are about 40 base pairs long and contain the instructions for the computation, and input molecules, which contain strings of six bases that represent the problem to be solved. A computation happens when these two types of molecules interact. Each has a sticky end, meaning one strand of the double helix is longer than the other, exposing a sequence of bases that are not paired.
When the sticky end of an input molecule bumps into a software molecule that has a sticky end that fits, the bases of the two sticky ends join together, and an enzyme present in the solution seals them together. The molecule is then cut in a different place by a second enzyme, exposing another sticky end so that a second step can take place. The number of steps depends on the number of computations coded into the software DNA strand.
For example, to answer the question 'does the sequence of letters "bab" contain an even number of b's?' the input DNA molecules would represent each of the three letters using a segment of DNA six bases long. The software molecules would contain a logical series of steps for determining whether the input has an even number of b's.
When the computation ends, the remaining sticky ends connect to one of two marker molecules of different lengths. One of the marker molecules contains a sequence of bases that will stick to the computation molecules' sticky end if the answer turned out to be even, and the other contains the correct sequences if the answer turned out to be odd.
Once the microscopic reshuffling of DNA bases is done, the researchers read the answer using gel electrophoresis, a process that involves putting DNA on a gel, and passing electrical current through electrodes attached to the gel. "[You] place the DNA near the minus electrode. The DNA travel slowly inside the gel towards the plus electrode at a speed that is a function of the molecule size. If you stop the process at the right time, you have a spread of the molecules according to... length," which makes it apparent which marker molecule has connected to the computation molecule, Shapiro said.
The researchers' DNA computer can run 735 sample programs made up of sequences of the eight basic operations that are possible using six-base input strings.
Previous experiments that used DNA to compute had to be designed to solve a single problem, although some could handle varied inputs, said Nadrian Seeman, a chemistry professor at New York University. "Here, several different questions can be asked of the same system," he said. The work could lead to more generally programmable approaches to DNA-based computation, he added.
In the researchers' experiments, the DNA took nearly 17 minutes to carry out each operation; eventually DNA will probably be able to carry out an operation in a few seconds, said Shapiro.
Compared to silicon computer chips, which compute in millionths or billionths of a second, this would still be very slow, but because millions or trillions of DNA molecules could compute in parallel, this type of DNA computer could handle very large problems; more important, it works in a test tube.
The work is "different from most experimental work on DNA computers in that we do not attempt to compete with silicon computers by solving difficult problems faster. The potential is... operation in a biochemical environment," said Shapiro.
The method is "an ingenious construction that provides the capability of a basic sort of computation at the molecular scale," said John Reif, a computer science professor at Duke University. It is not new to use the molecular configuration of DNA to store the result of a computation, but this work provides a very general mechanism for using that result to then compute a further result, he said.
This type of work could eventually lead to molecular computers that could be used to control molecular processes like complex assemblies and molecular robotic devices, Reif said.
The method could be used to screen libraries of DNA in five to ten years, and could eventually be used to carry out more complicated tasks like detecting DNA anomalies and synthesizing drugs to fix them, Shapiro said, adding that this use is decades away.
Shapiro's research colleagues were Yaakov Benenson, Tamar Paz-Elizur, Rivka Adar and Zvi Livneh from the Weizmann Institute, and Ehud Keinan from the Weizmann Institute and the Scripps Research Institute. They published the research in the November 22, 2001 issue of Nature. The research was funded by the Weizmann Institute.
Timeline: 5-10 years, several decades
Funding: University
TRN Categories: Biological, Chemical, DNA and Molecular Computing
Story Type: News
Related Elements: Technical paper, "Programmable and Autonomous Computing Machine Made of Biomolecules," Nature, November 22, 2001.
Advertisements:
November 28, 2001
Page One
Programmable DNA debuts
Device would boost quantum messages
Virtual computers reconfigure on the fly
Software sorts video soundtracks
Bigger disks won't hit quantum barrier
News:
Research News Roundup
Research Watch blog
Features:
View from the High Ground Q&A
How It Works
RSS Feeds:
News
Ad links:
Buy an ad link
| Advertisements:
|
 |
Ad links: Clear History
Buy an ad link
|
TRN
Newswire and Headline Feeds for Web sites
|
© Copyright Technology Research News, LLC 2000-2006. All rights reserved.