
Altered
protein orders metal bits
By
Eric Smalley,
Technology Research NewsGenetic engineering usually means experimental drugs, altered food crops and the specter of a new race of superhumans.
Researchers from NASA, the SETI Institute and Argonne National Laboratory have genetically modified a bacteria that lives in geothermal hot springs in order to make a microscopic scaffolding that produces a high-tech material.
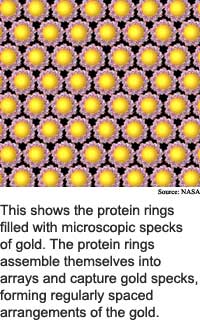
The altered bacteria gene produces a protein that automatically constructs orderly arrays of microscopic bits of gold or zinc. The microscopic bits of metal can serve as quantum dots, which trap one or a few electrons, and can be used in electronics.
Nanoscale arrays of quantum dots could be used to make data storage devices that hold enormous amounts of information; quantum computers that use quantum dots to compute, and materials that channel light waves in optical computers and communications devices.
The technique uses biotechnology and chemistry in place of conventional chipmaking processes. "The concept of using self-assembling biomolecules for materials science is not new," said Andrew McMillan, a research scientist at NASA's Ames Research Center. What is new is "we engineered [specific functions] into a protein that self-assembles into flat crystals," he said.
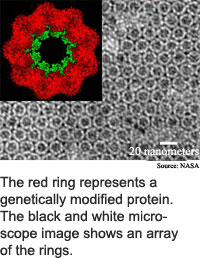
Certain portions of the protein bind to certain metals. When the protein tubes arrange themselves in a lattice, or crystal-like, structure, the result is uniform placement of nanoscale bits of metal, said McMillan.
The researchers used a protein from Sulfolobus shibatae, an extremophile organism that can grow in temperatures as hot as 85 degrees Celsius. The bacteria's ability to withstand high temperatures means the protein, called chaperonin, is particularly stable.
Chaperonins are essential proteins that exist in nearly all organisms, said McMillan. Their predominant function is thought to be facilitating protein folding inside cells, he said. Proteins fold to change shape at the molecular level, which allows them to carry out specific life processes.
Chaperonins are made of 14, 16 or 18 protein subunits arranged in a pair of stacked rings. "Our work takes advantage of the... characteristic barrel shape," said McMillan. The rings are 16 to 18 nanometers high by 15 to 17 nanometers in diameter. A nanometer is about the width of 10 hydrogen atoms.
The researchers used one of the three types of proteins that make up the chaperonin subunits. They altered the Sulfolobus genes to produce a protein with a slightly different structure, then inserted the altered genes into common E. coli bacteria, which manufactured large amounts of the modified protein. They heated the E. coli to 85 degrees Celsius to destroy the E. coli and its own proteins, leaving behind the engineered Sulfolobus protein.
The researchers engineered two variants of the protein. To get one variant, they removed a portion of the gene that causes bits of protein to partially block the openings at the ends of the chaperonin. The partially blocked variant had an opening of 3 nanometers and the unblocked variant had an opening of 9 nanometers.
They also mutated the genetic code of both variants to produce cysteine residues on the chaperonins' openings. Cysteine, an amino acid, acts like glue to bind a bit of gold or zinc to each opening like sticking a ball to the end of a tube.
The modified chaperonins did not lose their inherent stability or ability to self-assemble, said McMillan. "We were able to alter the protein without destroying its ability to form interesting structures," he said.
The researchers crystallized the altered chaperonins into flat, hexagonally packed templates, said McMillan. These chaperonin crystals arranged 5-nanometer quantum dots into arrays when the researchers used the variant with the 3-nanometer opening, while the the 9-nanometer chaperonin arranged 10-nanometer quantum dots.
The researchers' work expands the rapidly growing field of using biomolecules as nanoscale scaffolding to organize inorganic nanocrystals, said Shuguang Zhang, a principal research scientist and associate director of the Center for Biomedical Engineering at the Massachusetts Institute of Technology. The researchers used a well-understood protein complex that can assemble itself into useful structures, he said.
Proteins are particularly useful because researchers can modify their structures in precise locations without significantly altering their folding behavior, said Zhang. "This tailor-made approach will have tremendous impact on the growth of nanotechnology and nanobiotechnology," he said. "However, much effort is still needed to reduce the high cost of production and [improve the] stability of proteins in their complexes," said Zhang.
"I think the basic idea is neat, but the end result was not all that impressive," said Gerard Wong, an assistant professor of materials science and engineering at the University of Illinois at Urbana Champaign. "You have to compare what they wind up making with what people have already accomplished using block copolymer lithography and other competing technologies," he said.
Block copolymer lithography uses ultraviolet light to alter the structure of a mix of two polymers so that they form thin plastic films with closely packed arrays of nanoscale holes. The films can serve as templates for making data storage media or optical materials.
Protein-guided nanostructures could be used in practical applications in two to five years, said Jonathan D. Trent, a research scientist at NASA Ames Research Center. Sony Corporation and Matsushita Electric Industrial Co., Ltd. are pursuing protein-based techniques for manufacturing memory, he noted.
The researchers' next step is to see what other kinds of particles they can array using their protein scaffolding, said McMillan. "We hope to build on this model of self-assembling and see what we can push it to do," he said.
One challenge to using the technique for practical applications is determining whether and how to remove the protein templates, which could contaminate devices, after the arrays are formed, said McMillan.
McMillan and Trent's research colleagues were Chad D. Paavola of NASA's Ames Research Center, Jeanie Howard and Suzanne L. Chan of the SETI Institute, and Nestor J. Zaluzec of Argonne National Laboratory. They published the research in the November 25, 2002 issue of the journal Nature Materials. The research was funded by NASA, the U.S. Department of Energy (DoE) and the Defense Advanced Research Projects Agency (DARPA).
Timeline: 2-5 years
Funding: Government
TRN Categories: Biotechnology, Materials Science and Engineering, Nanotechnology
Story Type: News
Related Elements: Technical paper, "Ordered Nanoparticle Arrays Formed on Engineered Chaperonin Protein Templates," Nature Materials, November 25, 2002
Advertisements:
January 1/8, 2003
Page One
Interface gets the point
Altered protein orders metal bits
Hubs increase Net risk
Electron pairs power quantum plan
Aligned fields could speed storage
News:
Research News Roundup
Research Watch blog
Features:
View from the High Ground Q&A
How It Works
RSS Feeds:
News
Ad links:
Buy an ad link
| Advertisements:
|
 |
Ad links: Clear History
Buy an ad link
|
TRN
Newswire and Headline Feeds for Web sites
|
© Copyright Technology Research News, LLC 2000-2006. All rights reserved.